In this template Rapporter will present you Factor Analysis.
Factor Analysis is applied as a data reduction or structure detection method. There are two main applications of it: reducing the number of variables and detecting structure in the relationships between variables, thus explore latent structure behind the data, classify variables.
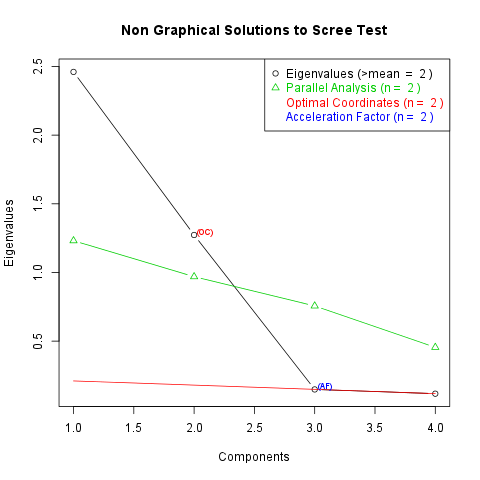
You can find the eigenvalues of the possible factors in the following table (2 factors were produced as you set):
| Factor Number | Eigenvalues |
|---|---|
| 1 | 2.461 |
| 2 | 1.273 |
| 3 | 0.1485 |
| 4 | 0.1176 |
At the next step let's check the factor loadings. They mean that how deep the impact of a factor for the variables. We emphasized the cells when the explained is higher than 30% of the whole variance.
| ML1 | ML2 | |
|---|---|---|
| carb | 0.8723 | 0.3756 |
| gear | -0.08811 | 0.9343 |
| mpg | -0.8197 | 0.4368 |
| cyl | 0.7988 | -0.452 |
So it can be said that
From them in the cases of the ML1's impact on mpg and ML2's impact on cyl, we can say they are negative effects.
At last but not least let us say some words about the not explained part of the variables. There are two statistics which help us quantifying this concept: Communality and Uniqueness. They are in a really strong relationship, because Uniqueness is the variability of a variable minus its Communality. The first table contains the Uniqunesses, the second the Communalities of the variables:
| Uniqunesses | Communalities | |
|---|---|---|
| carb | 0.09805 | 0.902 |
| gear | 0.1193 | 0.8807 |
| mpg | 0.1374 | 0.8626 |
| cyl | 0.1576 | 0.8424 |
We can see from the table that variable cyl has the highest Uniqueness, so could be explained the least by the factors and variable carb variance's was explained the most, because it has the lowest Uniqueness. From the communalities we can draw the same conclusion.
In this template Rapporter will present you Factor Analysis.
Factor Analysis is applied as a data reduction or structure detection method. There are two main applications of it: reducing the number of variables and detecting structure in the relationships between variables, thus explore latent structure behind the data, classify variables.
You can find the eigenvalues of the possible factors in the following table (3 factors were produced as you set):
| Factor Number | Eigenvalues |
|---|---|
| 1 | 2.461 |
| 2 | 1.273 |
| 3 | 0.1485 |
| 4 | 0.1176 |
At the next step let's check the factor loadings. They mean that how deep the impact of a factor for the variables. We emphasized the cells when the explained is higher than 30% of the whole variance.
| ML1 | ML2 | ML3 | |
|---|---|---|---|
| carb | 0.8771 | 0.3502 | -0.001772 |
| gear | -0.0595 | 0.9317 | -0.005685 |
| mpg | -0.8132 | 0.4641 | 0.0972 |
| cyl | 0.7917 | -0.4774 | 0.1361 |
So it can be said that
From them in the cases of the ML1's impact on mpg and ML2's impact on cyl, we can say they are negative effects.
At last but not least let us say some words about the not explained part of the variables. There are two statistics which help us quantifying this concept: Communality and Uniqueness. They are in a really strong relationship, because Uniqueness is the variability of a variable minus its Communality. The first table contains the Uniqunesses, the second the Communalities of the variables:
| Uniqunesses | Communalities | |
|---|---|---|
| carb | 0.1081 | 0.8919 |
| gear | 0.1283 | 0.8717 |
| mpg | 0.1138 | 0.8862 |
| cyl | 0.1268 | 0.8732 |
We can see from the table that variable gear has the highest Uniqueness, so could be explained the least by the factors and variable carb variance's was explained the most, because it has the lowest Uniqueness. From the communalities we can draw the same conclusion.
In this template Rapporter will present you Factor Analysis.
Your request cannot be implemented, because there are the same number of variables (5) like the number of the requested factors (5) . Please set the number of the factors to maximum 4 with the same number of the variables or extend the number of those variables at least to 6.
This report was generated with R (3.0.1) and rapport (0.51) in 1.479 sec on x86_64-unknown-linux-gnu platform.
